New publication for Alicia Russell; Genome mining strategies for ribosomally synthesised and post-translationally modified peptides.
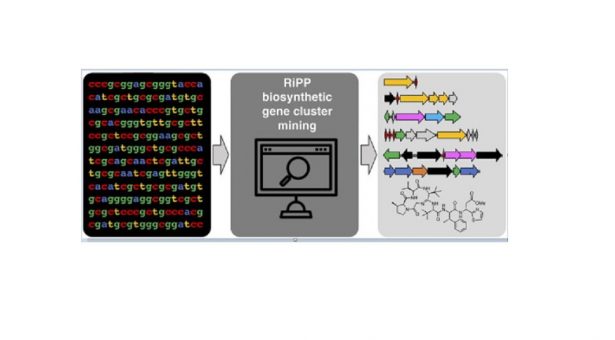
With her supervisor Andy Truman, NRPDTP student Alicia Russell wrote a review article about genome mining tools for RiPP natural products, which has been published in the Computational and Structural Biotechnology Journal.
RiPPs (ribosomally synthesised and post-translationally modified peptides) are a class of natural product that are structurally diverse and display a range of important biological activities such as antimicrobial and anticancer. It’s therefore really interesting and important to try to discover more of these compounds. Alicia’s PhD research focuses on the discovery of RiPP natural products from bacteria.
A common approach to natural products discovery is genome mining, which involves the automatic identification of biosynthetic genes from genome sequences. RiPPs have traditionally been particularly difficult to identify by genome mining because of their biosynthetic features, therefore lots of research groups have developed specific tools in recent years to try and identify RiPP gene clusters.
This review article summarises all of the currently available genome mining tools and databases for RiPPs, providing a single point of reference for researchers in our field to aid the discovery of novel compounds such as antibiotics. The discovery of new antimicrobials is particularly important at the moment because of antimicrobial resistance.
As part of the review article, Alicia also carried out a comparative analysis of different genome mining outputs, to show how they differ, and which ones are optimal for discovering and predicting certain types of compound.
The full article is available online
